- HUMAN PROTEIN ATLAS SECTIONS
- TISSUE
- BRAIN
- SINGLE CELL TYPE
- TISSUE CELL TYPE
- PATHOLOGY
- IMMUNE CELL
- BLOOD PROTEIN
- SUBCELLULAR
- CELL LINE
- METABOLIC
|
|
MethodMetabolic - Methods summarySummaryThe Metabolic Section contains information regarding the genes that encode for reactions in the human metabolic network. This network has been constructed in silico, and is the result of experiments whose data has been deposited in a wide range of databases and has references to around 2900 different articles and books. Key publicationsRobinson JL et al. (2020) "An atlas of human metabolism" Sci Signal. 12 (624): aaz1482 What can you learn from the Metabolic Section?Learn about:
How has the data been generated?The human metabolic network, Human-GEM, is the most recent development in a series of genome-scale metabolic models. Thus, Human-GEM integrates previously developed metabolic networks (Figure 1), reconciling information such as gene-reaction associations. Since the first public release in August 2018, Human-GEM has reached version 1.10.0 in its 39th release in September 2021.
Figure 1. The evolution of the human metabolic networks by publication year, with arrows indicating incorporation or reconciliation. What is presented in the section?The Metabolic Section presents gene information in 3 parts: general information, a metabolic summary table, and the pathway map. The general information includes the gene description, protein class, and other Human Protein Atlas information. In the metabolic summary table, the gene-reaction association is combined with reaction-subsystem associations and reaction-metabolite-compartment associations to deliver two key insights: what pathways/subsystems the gene contributes to, and in which cellular compartment(s) the associated reactions are expected to take place based on metabolite information. The size of the respective pathways are also included next to the number of reactions the gene catalyzes. The interactive part of the page is shown after clicking on a pathway in the metabolic summary table: the associated metabolic network map is shown, together with a gene-tissue heatmap that shows the expression of genes in that pathway among different tissues. |
The Project
The Human Protein Atlas

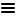 MENU
MENU